Table of Contents
The tryptophan operon in E.coli, required for synthesizing the amino acid tryptophan, is an example of a negatively controlled repressible operon. A negatively controlled operon is one that is inhibited by a regulatory protein called repressor. Tryptophan operon is a repressible system because it is inhibited when an effector molecule is present.
The structure of Tryptophan operon
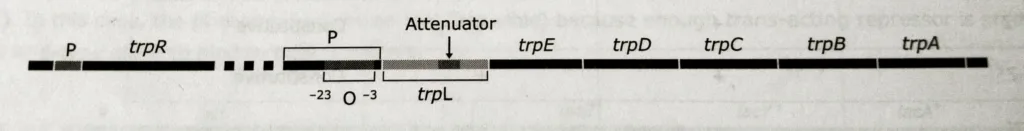
The tryptophan operon consists of five structural genes: trp E, trp D, trp C, trp B, trp that code for enzymes involved in the conversion of chorismic acid to tryptophan. The controlling site in trp operon lies next to trp E and consists of a promoter site (P), an overlapping operator site (O), and a leader region, trp L that codes for a leader peptide and an RNA attenuator. The leader peptide is involved in the attenuation mechanism.
Mechanism of action:
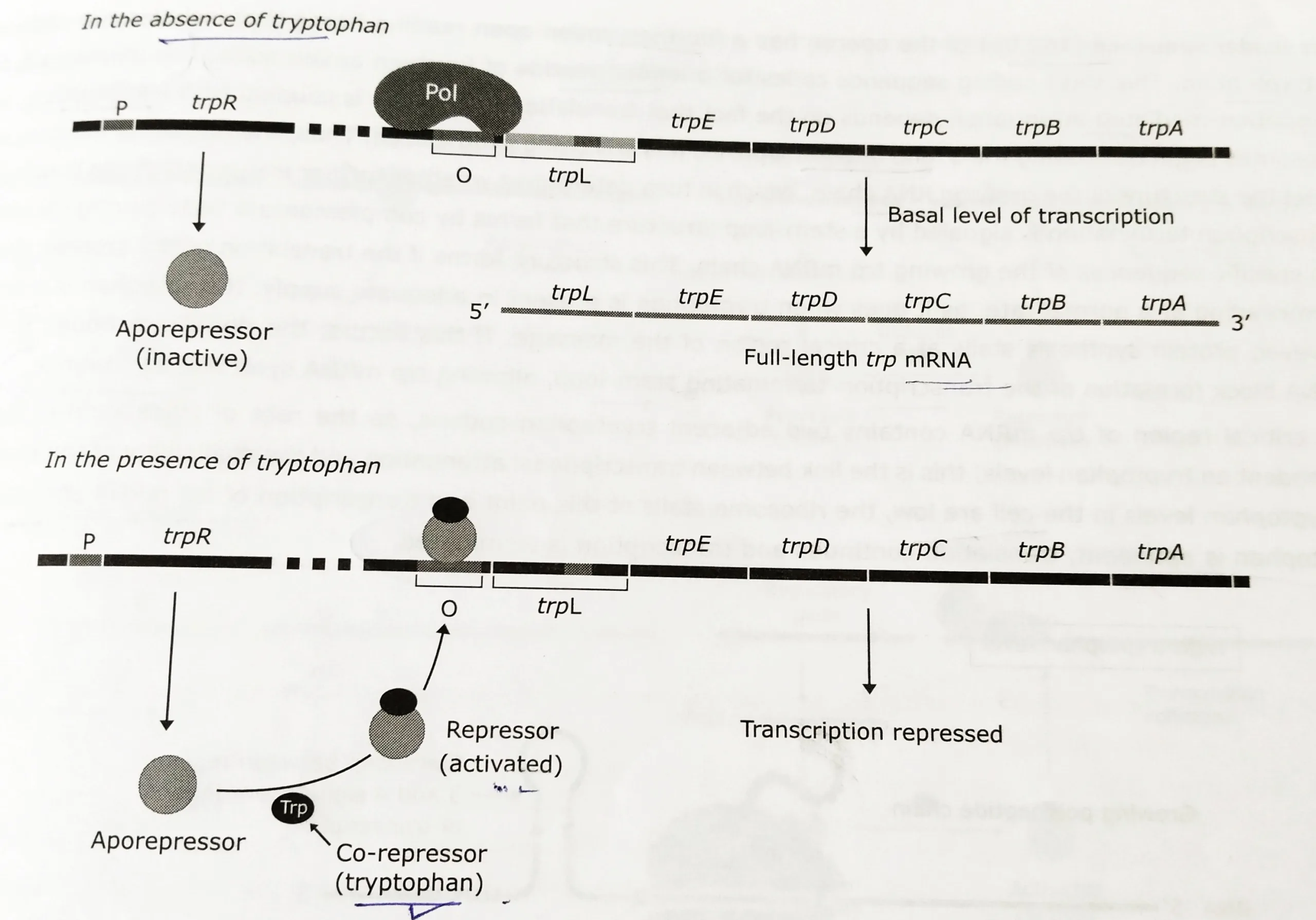
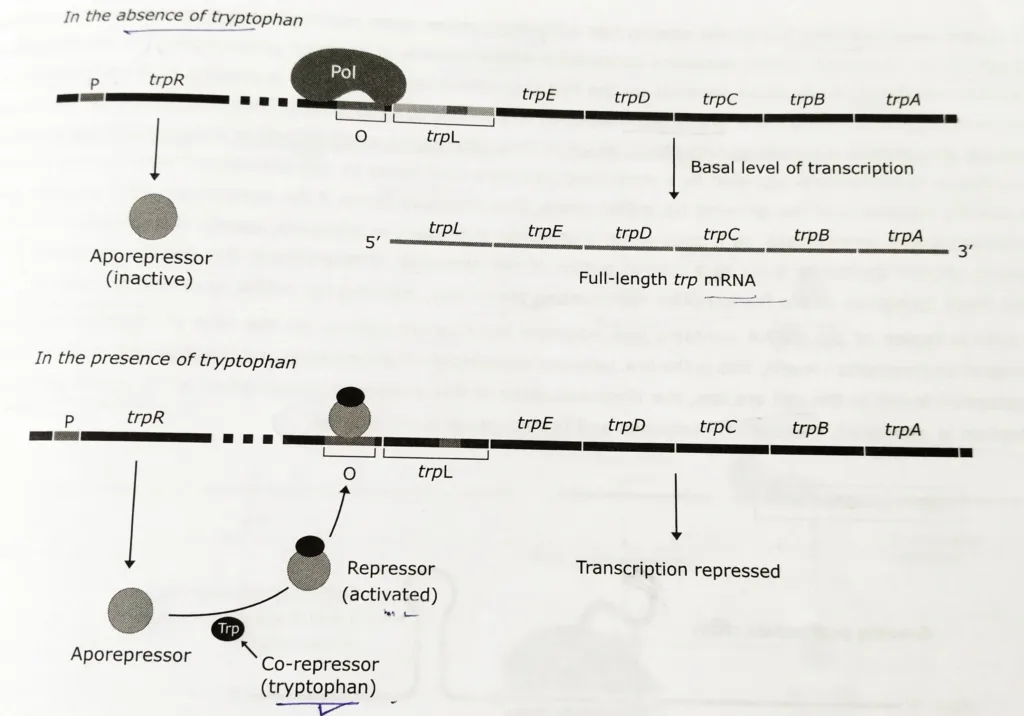
- In this repressible operon, the product of the trpR gene, the Trp repressor, is inactive by itself (called apo repressor); it doesn’t recognize the operator sequence of the trp operon. The repressor only becomes active when it combines with tryptophan.
- Hence, in the absence of tryptophan, the apo repressor (inactive regulatory protein) has no effect on the tryptophan operon. Consequently, the enzymes for synthesizing the amino acid are produced and the cell makes tryptophan.
- Tryptophan is referred to as the corepressor since it acts as an effector to convert the apo repressor into the active repressor.
- When tryptophan binds to the apo repressor, it induces a confirmational change that converts the regulatory protein into the active holorepressor.
- The holorepressor binds to the operator and blocks the attachment of RNA polymerase to a promoter. This shuts off the transcription of the structural genes.
Transcriptional attenuation
An additional mechanism termed transcriptional attenuation also negatively regulates the expression of many operons in bacteria by controlling the ability of RNA polymerase to continue elongation past specific sites. Transcriptional attenuation is carried out by two different processes mediated transcriptional attenuation and RNA-binding protein-mediated attenuation. They are explained below.
Translation mediated transcriptional attenuation
- In E. coli, provides an additional level of control that results in more stringent regulation that could be achieved by repression of transcription initiation alone. In trp operon, the site attenuation is located downstream of the transcription start site.
- If tryptophan is abundant, most transcription terminates at this site; only if tryptophan is scarce does transcription continue to yield functional trp mRNA.
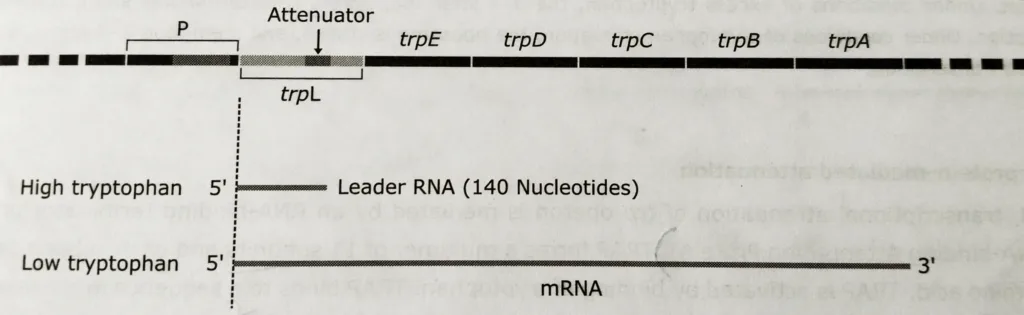
- The leader sequence (162 bp) of the operon has a fourteen codon open reading frame that includes two codons of tryptophan. This short coding sequence codes for a leader peptide of fourteen amino acids.
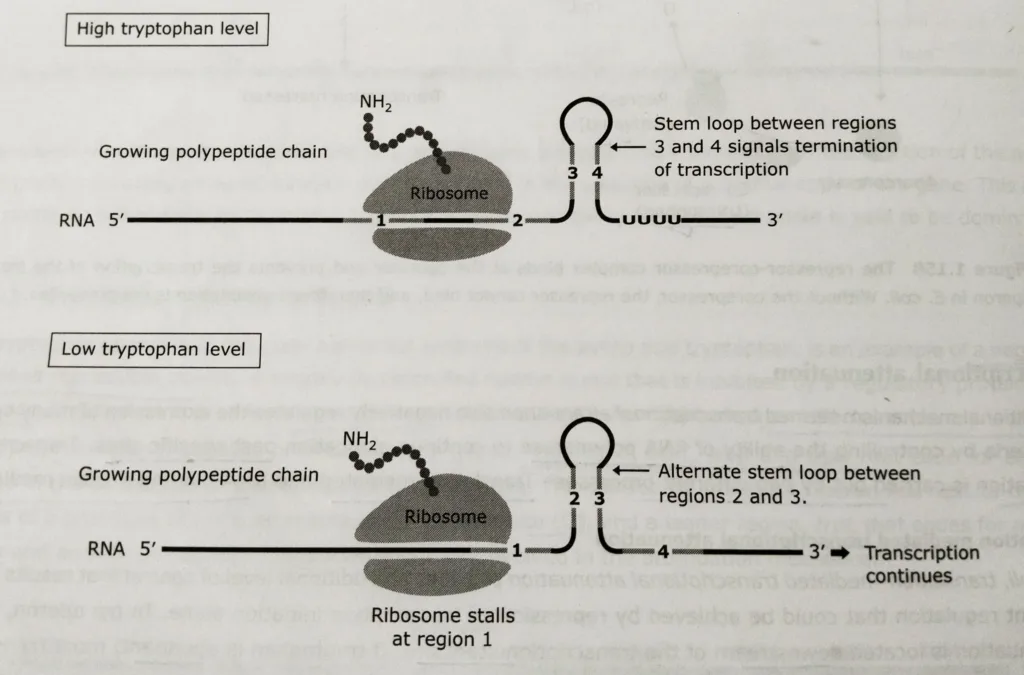
- The mechanism of translation -mediated attenuation depends on the fact that translation in bacteria is coupled with transcription, so ribosomes begin translating the 5′ end of a mRNA while it is still being synthesized.
- Thus, the rate of translation can affect the structure of the growing RNA chain, which in turn determines whether further transcription can continue.
- Transcription termination is signalled by a stem-loop structure that forms by complementary base pairing between two specific sequences of the growing trp mRNA chain.
- This structure forms if the translation of the growing chain is proceeding at a normal rate, as it does when tryptophan is present in adequate supply.
- If tryptophan is scarce, protein synthesis stalls at a critical region of the message. If this occurs, the ribosomes bound to the mRNA block formation of the transcription-terminating stem-loop, allowing trp mRNA synthesis to continue.
- The critical region of trp mRNA contains two adjacent tryptophan codons, so the rate of translation is highly dependent on tryptophan levels. This is the link between transcriptional attenuation and the availability of tryptophan.
- If the tryptophan levels in the cell are low, the ribosomes stalls at the point of transcription of trp mRNA continue. If the tryptophan is abundant, translation continues and the transcription is terminated.
RNA-binding protein-mediated attenuation
- In B. subtilis, transcriptional attenuation of tryptophan operon is mediated by an RNA-binding terminator protein called TRAP (Trp RNA-binding Attenuation protein).
- TRAP forms a multimer of 11 subunits mad each subunit binds a single tryptophan amino acid. TRAP is activated by binding of tryptophan. TRAP binds to a sequence in the leader segment of the transcript.
- Binding of TRAP causes the formation of the terminator hairpin. The termination of transcription then prevents the production of the tryptophan biosynthetic enzymes.
- In the absence of tryptophan, TRAP does not bind, and the mRNA adopts a structure that prevents the terminator hairpin from forming. Protein anti-TRAP controls the activity of TRAP, which binds to TRAP, and prevents it from repressing the tryptophan operon.
Other related notes:
- Nondisjunction and Aneuploidy: https://thebiologyislove.com/nondisjunction-and-aneuploidy/
- RAS-MAP kinase pathway: https://thebiologyislove.com/ras-map-kinase-pathway/
- Cell cycle checkpoints: https://thebiologyislove.com/cell-cycle-checkpoints/
Facebook link: https://www.facebook.com/share/p/1QCBBX8xPWiwQE7T/?mibextid=oFDknk
Instagram link: https://www.instagram.com/p/C7o7lOdJrR0/?igsh=dTBzOWNja2xvbXJ1