Table of Contents
Prophase 1 is the first phase (Substage) of Meiosis 1. This substage has five sequential phases- Leptotene, Zygotene, Pachytene, Diplotene, and Diakinesis. The most important event of the meiotic cell division is crossing over, which occurs here in Prophase 1. The substage Prophase 1 is explained step by step below. The mnemonic of the sub-stages of the Prophase 1 is provided here. Prophase 1 of meiosis 1 is different from the prophase of mitosis type of cell division.
Duplicated Homologs pair during Meiotic Prophase 1
- The gradual juxtaposition of homologs occurs during a prolonged period called meiotic prophase (or prophase 1), which can take hours in yeasts, days in mice, and weeks in higher plants.
- Like their mitotic counterparts, duplicated meiotic prophase chromosomes first appear as long threadlike structures in which the sister chromatids are so tightly attached together that they appear as one.
- It is during early prophase 1 that the homologs begin to associate along their length in a process called pairing, which, in some organisms at least, begins interactions between complementary DNA sequences (called pairing sites) in the two homologs.
- The homologs become more closely juxtaposed as prophase progresses, generating a four-chromatid structure called bivalent.
- In most species, homolog pairs are then locked together by homologous recombination: DNA double-strand breaks are formed at several locations in each sister chromatid, leading to large numbers in DNA recombination events between the homologs.
- Some of these events lead to reciprocal DNA exchanges called crossovers, where the DNA of a chromatid crosses over to become continuous with the DNA of a homologous chromatid.
Homolog pairing leads up to the formation of a Synaptonemal Complex
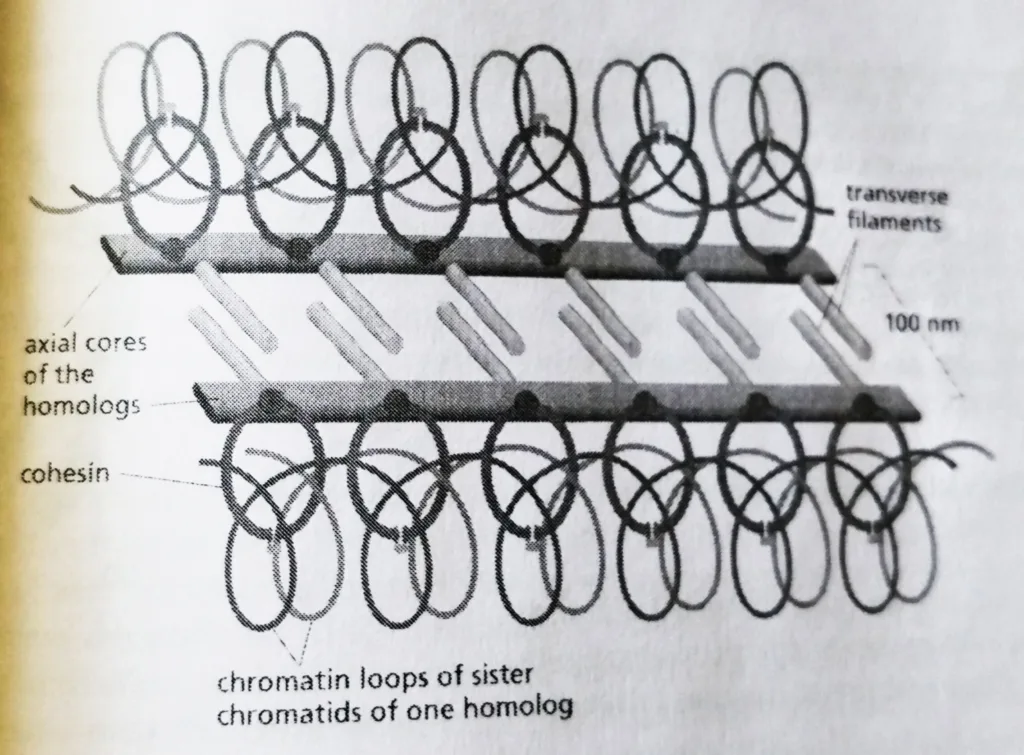
- The paired homologs are brought into close juxtaposition, with their structural axes (axial cores) about 400 nm apart, by a mechanism that depends in most species on the double-strand DNA breaks that occur in sister chromatids.
- One possibility is that the large protein machine called a recombination complex. The recombination complex assembles on a double-strand break in a chromatid, binds the matching DNA sequences in the nearby homolog and helps reel in this partner.
- This so-called presynaptic alignment of the homologs is followed by synapsis, in which the axial core of a homolog becomes tightly linked to the axial core of its partner by a closely packed array of transverse filaments to create a synaptonemal complex, which bridges the gap, now only 100 nm, between the homologs.
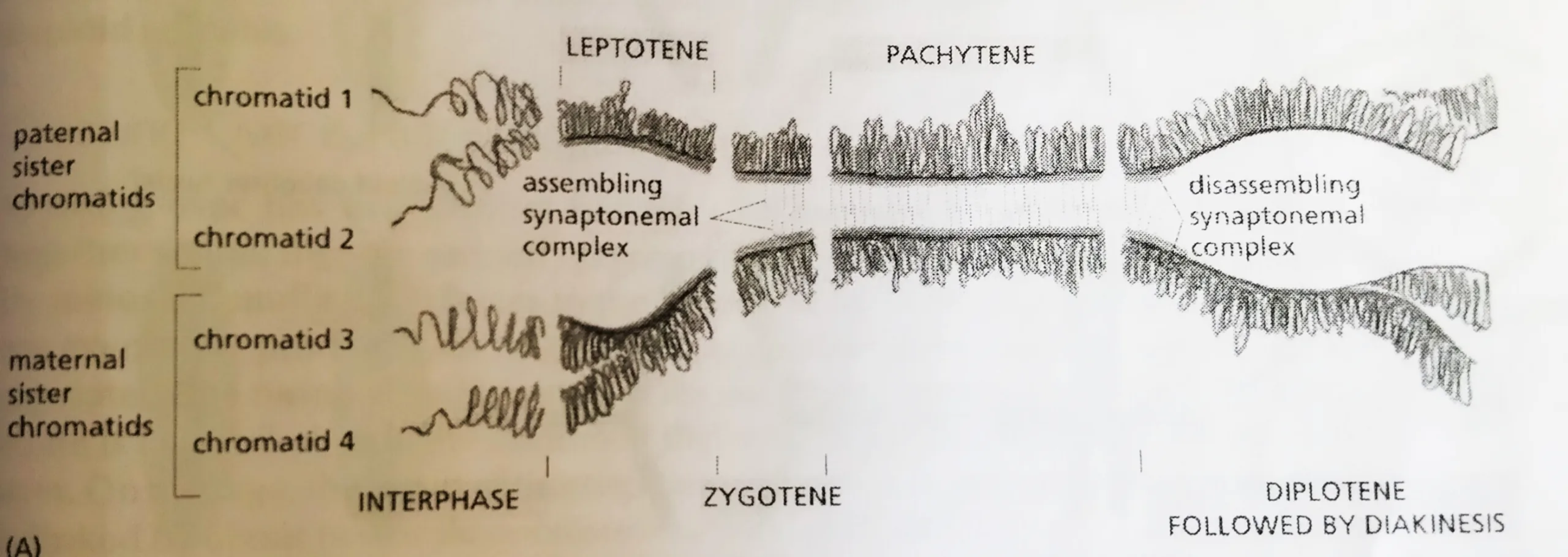
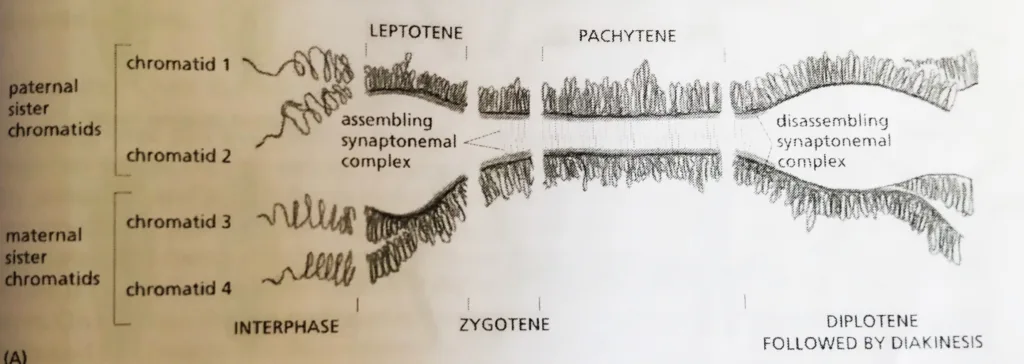
- The morphological changes that occur during homolog pairing are the basis for dividing meiotic prophase into five sequential stages– Leptotene, Zygotene, Pachytene, Diplotene, and Diakinesis.
- Prophase starts with leptotene, which homologs condense and pair and genetic recombination begins.
- At Zygotene, the synaptonemal complex begins to assemble at sites where the homologs are closely associated and recombination events are occurring.
- The assembly process is complete at Pachytene, and the homologs are synapsed along their entire lengths.
- The Pachytene stage can persist for days or longer, until desynapsis begins at Diplotene with the disassembly of the synaptonemal complexes and the concomitant condensation and shortening of the chromosomes.
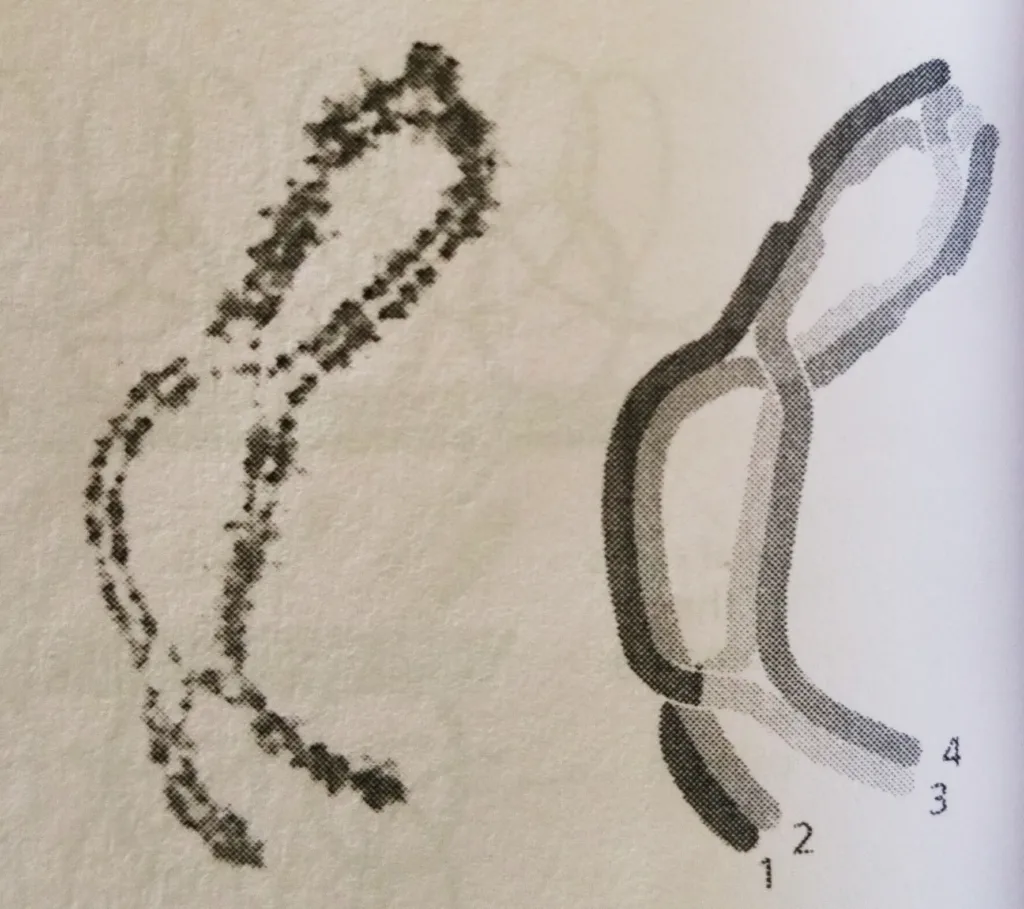
- It is only at this stage, after the complexes have disassembled, that the individual crossover events between non-sister chromatids can be observed as inter-homolog connections called chiasmata (singular chiasma), which now play a crucial part in holding the compact homologs together, The homologs are now ready to initiate the process of segregation.
Homolog pairing and crossing-over:
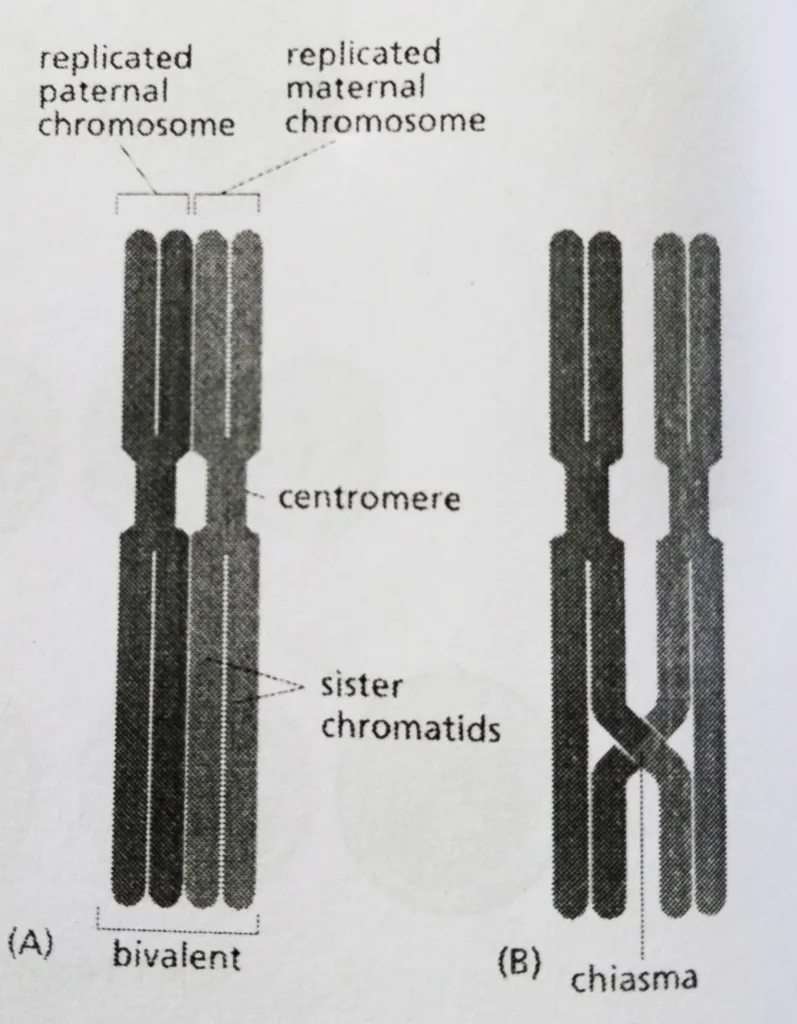
- The structure formed by two closely aligned duplicated homologs is called a bivalent.
- As in mitosis, the sister chromatids in each homolog are tightly connected along their entire lengths, as well as their centromeres.
- At this stage, the homologs are usually joined by a protein complex called as synaptonemal complex.
- A later-stage bivalent in which a single crossover has occurred between non-sister chromatids.
- It is only when the synaptonemal complex disassembles and the paired homologs separate a little at the end of prophase 1.
Schematic diagram of a synaptonemal complex:
Homolog Synapsis and desynapsis during the different stages of prophase 1:
Other related notes:
- Cell cycle checkpoints: https://thebiologyislove.com/cell-cycle-checkpoints/
- Useful numbers in cell culture: https://thebiologyislove.com/useful-numbers-for-cell-culture/
Facebook link: https://www.facebook.com/share/p/eheZ11mspxjc8imm/?mibextid=oFDknk
Instagram link: https://www.instagram.com/p/C6dddLdR0fm/?igsh=OGRyMHdiYWhlOXky