Table of Contents
Polyadenylation is one of the major methods of the processing of eukaryotic pre-mRNA after the capping mechanism.
Introduction to polyadenylation:
Virtually all eukaryotic mRNAs have a series of up to 250 adenosines at their 3′ ends called poly-A tail. A poly-A tail is added to the 3′ end of mRNAs and plays an important role in mRNA stability, nucleocytoplasmic export, and translation.
The poly-A tail is not specified by the DNA and is added to the transcript by a template-independent RNA polymerase called poly-A polymerase (PAP) after endo nucleolytic cleavage of the nascent RNA near the 3′ terminus. These two coupled reactions (cleavage and formation of poly-A tail), are collectively referred to as cleavage and poly-adenylation or, simply, polyadenylation.
The steps of polyadenylation:
A single processing complex undertakes both the cleavage and poly-adenylation and a signal is required.
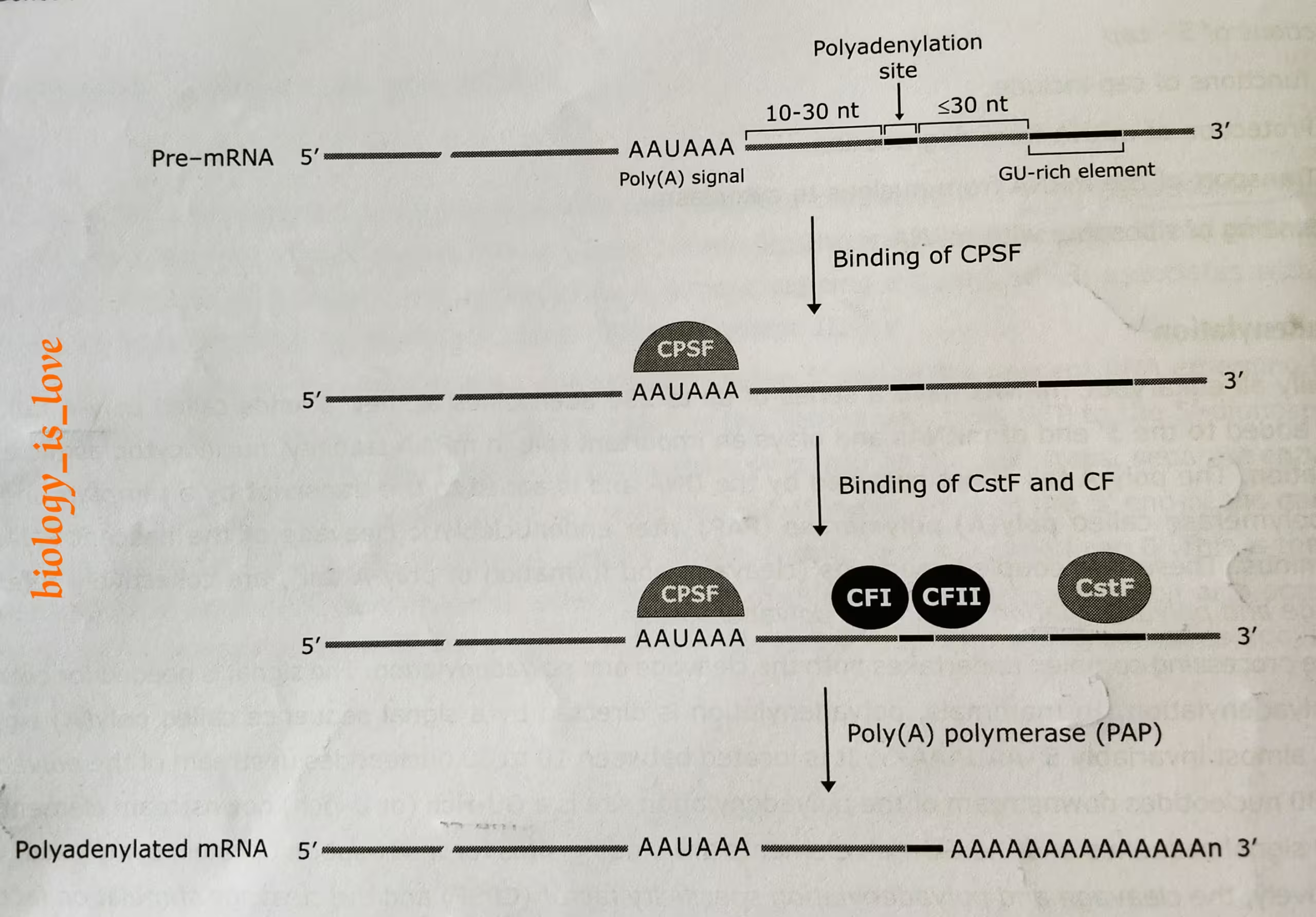
Step 1:
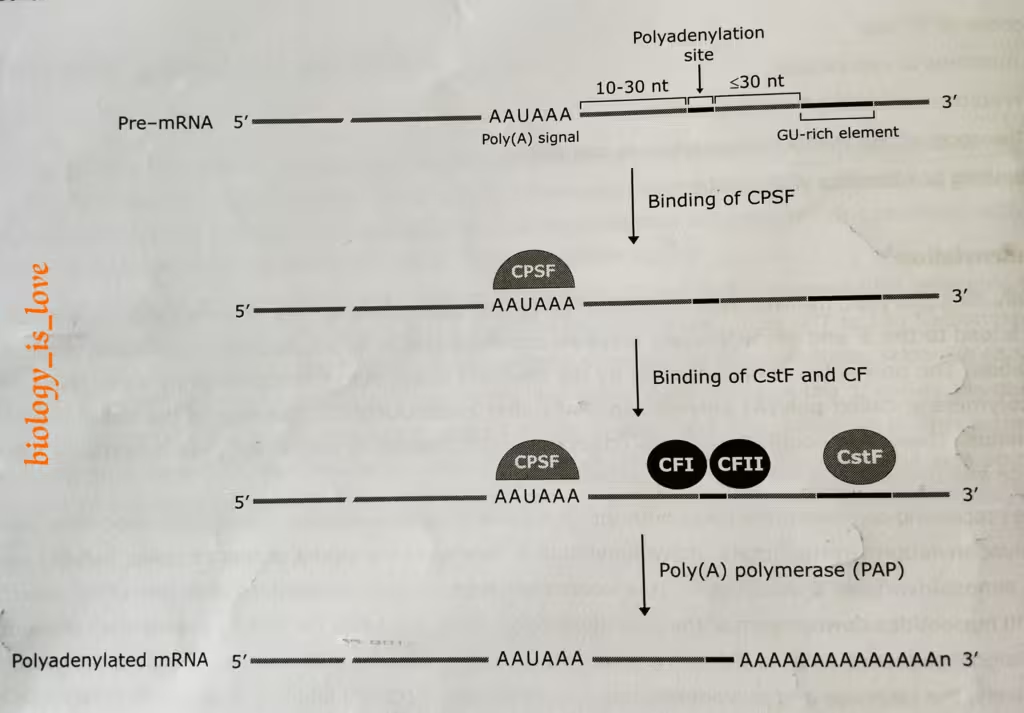
- In mammals, this process is directed by a signal sequence called poly-A signal in the mRNA, almost invariably 5′-AAUAAA-3′ (upstream element).
- It is located between 10 to 30 nucleotides upstream of the polyadenylation site.
- Almost equal and lesser than 30 nucleotides downstream of the polyadenylation site is a GU-rich (or U-rich) downstream element.
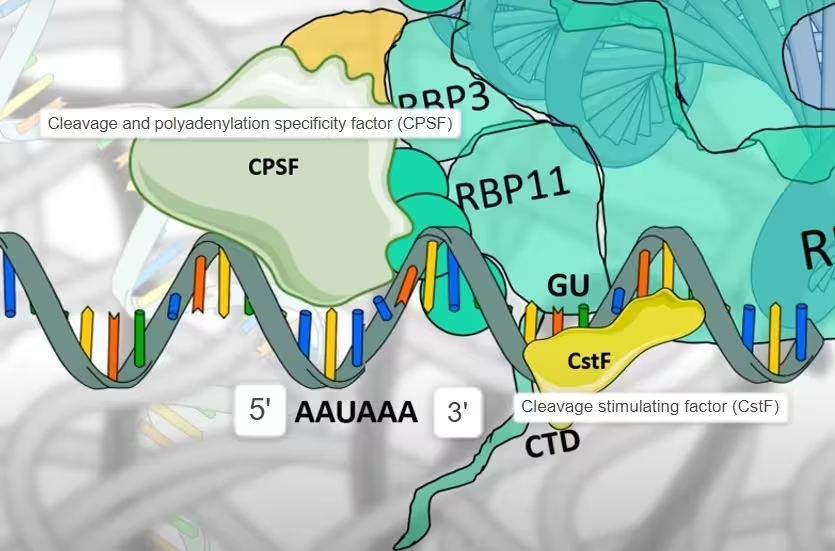
- Both the poly-A signal sequence and the GU rich elements are the binding sites for the multi-subunit protein complexes, which are which are respectively, the cleavage and poly-adenylation specificity factor (CPSF) and the cleavage stimulating factor (CstF).
The diagram, describes the above step 1:
Step 2:
- Generation of poly-A tail at 3′ terminus first requires cleavage factors- CFI and CF II which act as endonucleases to cleave the RNA at poly-adenylation site.
The diagram, describes the above step 2:
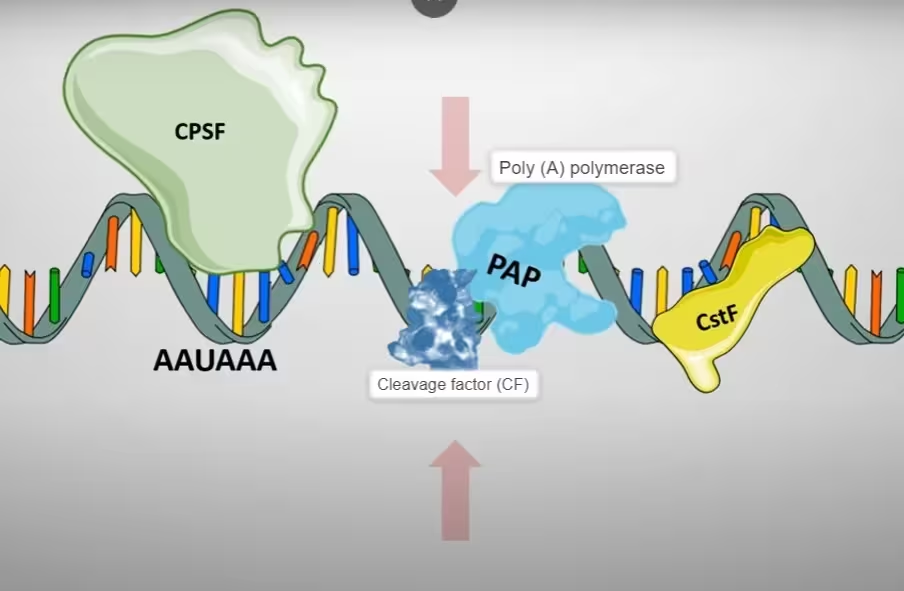
- Cleavage is followed by the poly-adenylation in a tightly coupled manner. The poly (A) polymerase (PAP) catalyzes the target reaction in template independent manner. Its substrate is ATP.
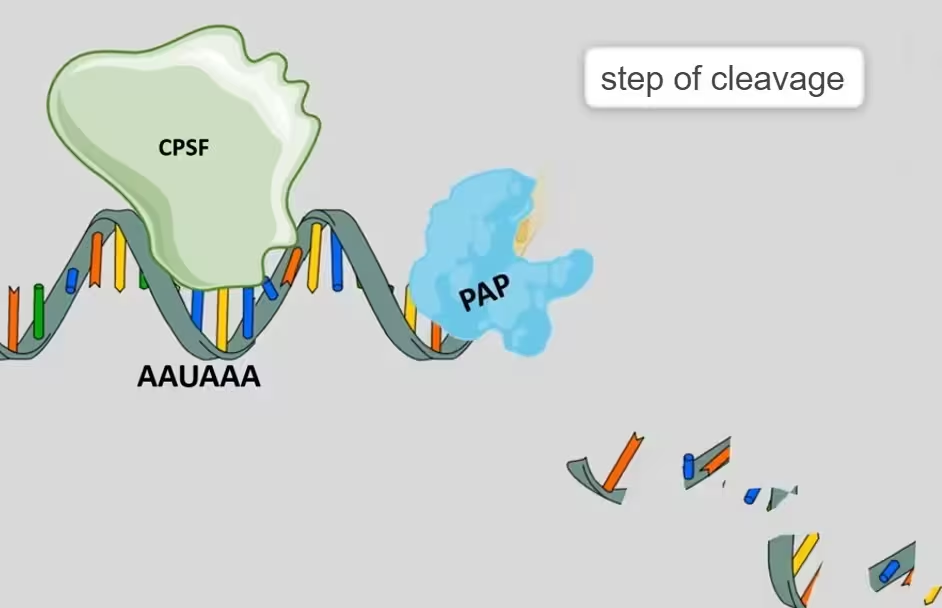
Step 3:
The reaction of poly-adenylation passes through two consecutive stages-
- A rather short oligo-A sequence (~10 residues) is added to the 3′ end.
- The oligo-A tail is extended to the full ~200 residue length.
The diagram, describes the above step 3:
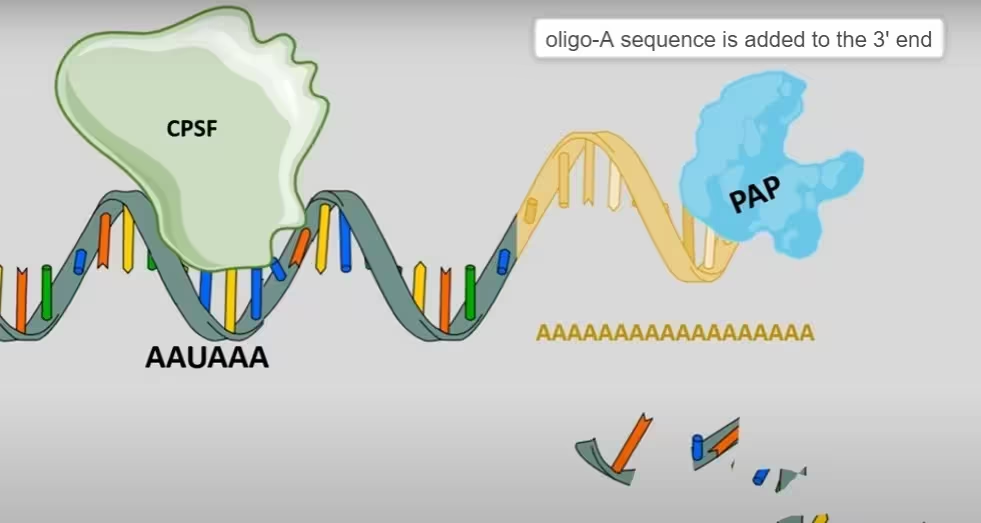
Step 4:
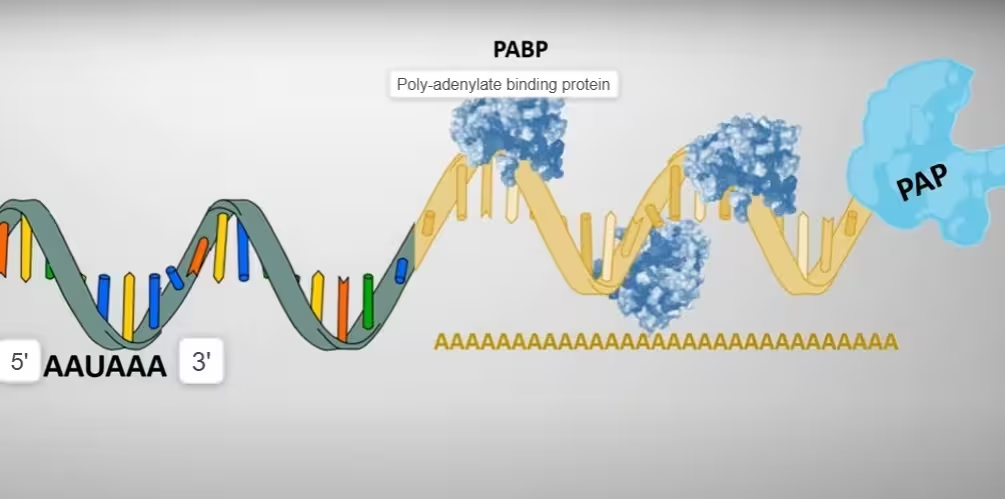
- This reaction needs another stimulatory factor that identifies the oligo-A tail.
- This additional factor includes poly-adenylate-binding protein (PABP), which helps the polymerase to add the adenosines.
- PABP influences the length of the poly-A tail that is formed and appears to play a role in the maintenance of the tail after synthesis.
The diagram, describes the above step 4:
The schematic representation of the process of polyadenylation:
Conclusion:
- This process also occurs in some mRNA in bacteria.
- In E. coli, this process in some mRNA leads to the formation of poly-A tail of 10-40 nucleotides.
- It is catalyzed by poly (A) polymerase associated with ribosomes.
- Here, poly-A tail acts as a binding site for the nucleases (PNPase and RNaseE) responsible for degradation of mRNA.
Other related notes:
- 5′ capping: https://thebiologyislove.com/5-capping/
- Alternative poly-adenylation: https://thebiologyislove.com/alternative-polyadenylation/
Facebook link: https://www.facebook.com/share/p/R7b2QRNv166p97aG/?mibextid=oFDknk
Instagram link: