Table of Contents
Base excision repair involves removal of a damaged nucleotide base, excision of a short piece of the polynucleotide and resynthesis with a DNA polymerase.
The objective of the Base Excision repair
It is used to repair minor damage like alkylation and deamination resulting from exposure to mutagenic agents.
Role of DNA glycosylase:
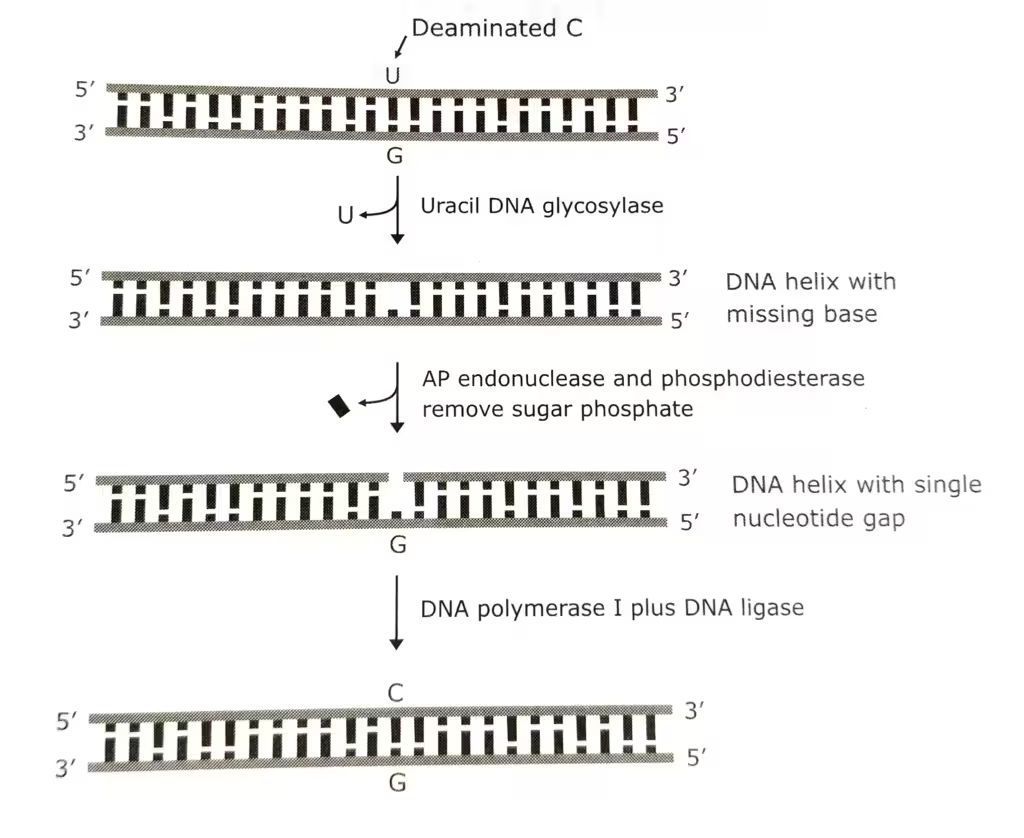
Enzyme DNA glycosylase initiates the repair process. A DNA glycosylase does not cleave phosphodiester bonds, instead, it cleaves the N-glycosidic bonds, liberating the altered base and forming an apurinic or an apyrimidinic site, both AP sites. The resulting AP site is then repaired by an AP endonuclease repair pathway.
The main mechanism: role of AP endonuclease:
- All cells have endonucleases that attack the sites left after the spontaneous loss of single purine or pyrimidine residues.
- The AP endonucleases are vital to the cell, because spontaneous depurination is a relatively frequent event.
- These enzymes introduce chain breaks by cleaving the phosphodiester bonds at AP sites.
- This bond cleavage initiates an excision-repair process with the help of three enzymes- an exonuclease, DNA polymerase I, and DNA ligase.
The schematic representation of Base excision repair mechanism:
Other related notes:
- Cell cycle checkpoint: https://thebiologyislove.com/cell-cycle-checkpoints/
- Nucleotide excision repair: https://thebiologyislove.com/nucleotide-excision-repair/
- Non homologous end joining:
Facebook link: https://www.facebook.com/share/p/YPes3eXQRbrtRuMU/?mibextid=oFDknk
Instagram link: https://www.instagram.com/reel/C8WuLMhpwM_/?igsh=NGJoZm5kNGNwYWVq