Table of Contents
Alternative splicing is a major type of RNA splicing method, occurs in mainly eukaryotic genes. The detailed mechanisms with examples are described here.
The mechanism- in brief:
During RNA splicing, the introns are precisely excised and the exons ligated together. The majority of nuclear pre-mRNA are spliced constitutively; that is, only one mature mRNA species is generated from a single pre-mRNA in all tissues.
In some cases, alternate 5′ and /or 3′ splice sites are used during splicing, resulting in the production of more than one mRNA species from single pre-mRNA. The production of different RNA products from a single product by changes in the usage of splicing junctions is described as alternative splicing.
This splicing process has been documented for many eukaryotic genes. The utilization of alternative 5′ and/or 3′ splice sites can result in structurally distinct mRNAs by either excluding potential exon sequences or incorporating otherwise noncoding intron sequences. For some pre-mRNAs, this process is a non regulated event. For others, the choice of alternative splice sites is regulated in a tissue-specific manner.
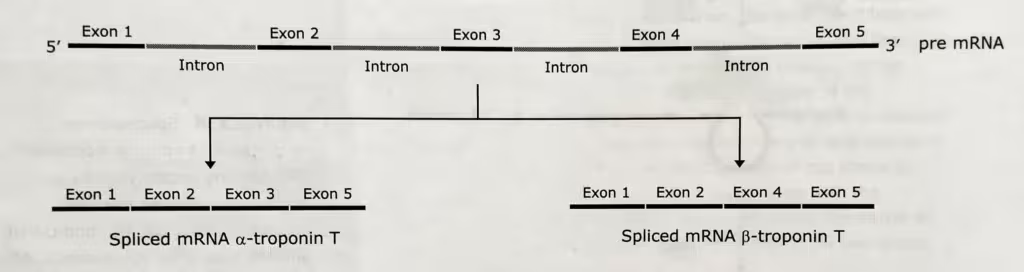
The alternative splicing in the Troponin T-gene’s transcript:
Troponin T-gene encoding 5-exons which generates two alternatively spliced mRNA formed as indicated in the above picture. One contains exons 1, 2, 3, and 5; the other contains 1, 2, 4, and 5.
The diagrammatic representation of the alternative-splicing in the Troponin gene:
The best example of alternative splicing:
Drosophila sex determination provides the best example of the regulated alternative splicing.
About Drosophila sex determinants:
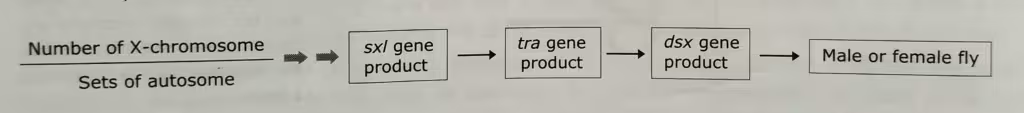
- In Drosophila, the primary signal for determining whether the fly develops as a male or female is the ratio of the number of X-chromosome and autosome set.
- Three crucial gene products are involved in transmitting information about the ratio to many other genes that specify male and female characteristics.
- The genes are called sex-lethal (sxl), transformer (tra) and doublesex (dsx).
- The function of these gene products is to transmit the information about the X-chromosome/autosome sets ratio to the many other genes that are involved in creating the sex-related phenotypes.
The word diagram of the sex determination in Drosophila:
The splicing mechanism:
- Regulated alternative splicing during expression of genes involved in sex determination in Drosophila.
- The first gene that undergoes alternative splicing is sxl, whose transcript contains an optional exon which results in an inactive version of Sxl protein.
- Functional sxl protein in female blocks the 3′ splice site in the first intron of the tra pre mRNA. This results in an mRNA that codes for a functional Tra Protein.
- In males, there is no functional sxl so that 3′ splice site is not blocked and a dysfunctional tra mRNA is produced. The resulting female specific Tra protein is again involved in alternative splicing.
- Tra protein interacts with splicing factors and binds within an exon of a third pre -mRNA (product of dsx gene). This results in female specific Dsx protein.
- The two versions of Dsx proteins are the primary determinants of Drosophila sex.
The diagrammatic representation of the above description:

Other related links:
- 5′ capping: https://thebiologyislove.com/5-capping/
- Polyadenylation: https://thebiologyislove.com/polyadenylation/
- Post-translational modifications of protein: https://thebiologyislove.com/post-translational-modifications-of-proteins-3-events/
Facebook link: https://www.facebook.com/share/p/V9R8p3Cpx4g641yn/?mibextid=oFDknk
Instagram link:

